pMeca Plasmid
pMECA is a cloning vector which contains 44 unique restriction sites, including 9 rare-cutters, all within the 230-bp polylinker. This allows for the linking together of up to eighteen different gene cassettes prior to transformation into the target organism.
Highlights:
- Highly compatible - use with plants, vertebrates, fungi, and bacteria
- Multiple cloning site (MCS) contains 44 unique restriction sites, including 9 rare-cutters
- Allows for concomitant use of multiple gene cassettes
- Traditional blue/white screening identifies bacterial colonies with inserts cloned into the vector
- Bacterial colonies whose vectors contain inserts grow much faster than those without, permitting the size of the bacterial colonies to be used to determine those which contain inserts
- Strains that have been transformed with the vector: DH5alpha, DH5alpha (MCR), Top10, JM109, HB101
Plasmid vectors form one of the central cores of molecular biology by allowing the manipulation of natural and synthetic DNA molecules. The exploitation of current vectors, which are of limited capabilities, has led to the need for new and inventive vectors which increase the maneuverability of DNA. The pMeca plasmid is a computer designed synthetic DNA vector system that allows for the ability to transform an organism with multiple genes making it easier to stack “transgenes” together or engineer novel metabolic pathways. The plasmid is ideal for large scale applications, speeding up the generation of transgenes and simplifying discovery and development work, with a minimal number of steps.
From the laboratory of Wayne A. Parrott, PhD, University of Georgia.
pMECA is a cloning vector which contains 44 unique restriction sites, including 9 rare-cutters, all within the 230-bp polylinker. This allows for the linking together of up to eighteen different gene cassettes prior to transformation into the target organism.
Highlights:
- Highly compatible - use with plants, vertebrates, fungi, and bacteria
- Multiple cloning site (MCS) contains 44 unique restriction sites, including 9 rare-cutters
- Allows for concomitant use of multiple gene cassettes
- Traditional blue/white screening identifies bacterial colonies with inserts cloned into the vector
- Bacterial colonies whose vectors contain inserts grow much faster than those without, permitting the size of the bacterial colonies to be used to determine those which contain inserts
- Strains that have been transformed with the vector: DH5alpha, DH5alpha (MCR), Top10, JM109, HB101
Plasmid vectors form one of the central cores of molecular biology by allowing the manipulation of natural and synthetic DNA molecules. The exploitation of current vectors, which are of limited capabilities, has led to the need for new and inventive vectors which increase the maneuverability of DNA. The pMeca plasmid is a computer designed synthetic DNA vector system that allows for the ability to transform an organism with multiple genes making it easier to stack “transgenes” together or engineer novel metabolic pathways. The plasmid is ideal for large scale applications, speeding up the generation of transgenes and simplifying discovery and development work, with a minimal number of steps.
From the laboratory of Wayne A. Parrott, PhD, University of Georgia.
| Product Type: | Plasmid |
| Name: | pMeca |
| Accession ID: | AF017063 |
| Antibiotic Resistance: | Amp |
| Fusion Tag(s): | lacZ |
| Tested Applications: | Development of novel transgenic crops and transgenic bacteria for multiple uses in the biotech and pharmaceutical industries |
| Grow in E. coli at 37 C: | Yes |
| Selectable markers: | Blue/white |
| Cloning Site 5': | EcoRI |
| Cloning Site 3': | HindIII |
| Vector Backbone and Size: | 2860 bp |
| High or low copy: | High |
| Storage: | -20C |
| Shipped: | Ambient temperature, spotted filter paper |
![]() pMeca Plasmid - Suggested Guidelines
pMeca Plasmid - Suggested Guidelines
It is suggested to first plate the transformed cells on LB/Amp with IPTG and X-gal. Once familar with the growth characteristics, then remove the use of IPTG and X-gal. Bacteria transformed with the vector will also take longer to appear blue. One trick is to place the plates in the refrigerator (after growth at 37C) for several hours to better see the blue phenotype. Also, it is easy to pick based on size, so it is not necessary to wait a long time for the small colonies to grow or become blue; just select the large colonies on the plate after 16 hrs of growth. While it is not necessary to use chloramphenicol for the production of plasmid, some may find it necessary.
Strain DH10B has been reported to not be transformable with pMECA. That appears to not be the case once pMECA has an insert. This can be an advantage in that only pMECA clones containing an insert grow well in this strain. It is recommended that the vector first be transformed into DH5alpha to grow up stock plasmid. Then this stock can be used to clone into and transformed into DH10B. In theory only the clones that appear on the plate should be recombinant.
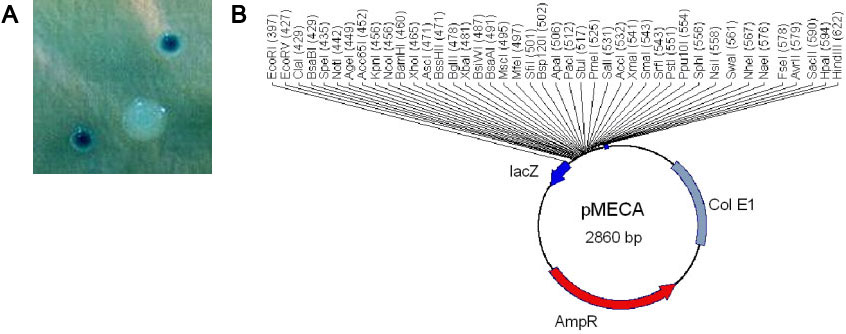
(A) pMECA colonies with (large and white) and without (small and blue) inserts. (B) pMeca Plasmid Map. The multiple cloning site is revealed as an EcoRI/HindIII fusion with the lacZ gene. Unique restriction sites are shown, and the same numbers are used for neoschizomers (isoschizomers that cut at different sites within the same sequence).
- Thomson, JM, and Parrott WA. pMECA: A cloning plasmid with 44 unique restriction sites that allows selection of recombinants based on colony size. 1998. BioTechniques. 24:922-927.
- Parrott, WA. "Transformation Vector System." US Patent Number 6,096,523; Aug 1, 2000. Pending applications in several jurisdictions University of Georgia Research Foundation, Inc.
pMeca Plasmid characterization
- Hartwich H, Nothwang HG. An easy and versatile 2-step protocol for targeted modification and subcloning of DNA from bacterial artificial chromosomes using non-commercial plasmids. BMC Res Notes. 2012 Mar 20;5:156. doi: 10.1186/1756-0500-5-156
- Huang YH, Cozzitorto JA, Richards NG, Eltoukhy AA, Yeo CJ, Langer R, Anderson DG, Brody JR, Sawicki JA. CanScript, an 18-Base pair DNA sequence, boosts tumor cell-specific promoter activity. Cancer Biol Ther. 2010 Nov 1;10(9):878-84. doi: 10.4161/cbt.10.9.13234. Epub 2010 Nov 1.
- Garg RP, Parry RJ. Regulation of valanimycin biosynthesis in Streptomyces viridifaciens: characterization of VlmI as a Streptomyces antibiotic regulatory protein (SARP). Microbiology. 2010 Feb;156(Pt 2):472-83. doi: 10.1099/mic.0.033167-0. Epub 2009 Nov 5.
- Colby JK, Klein RD, McArthur MJ, Conti CJ, Kiguchi K, Kawamoto T, Riggs PK, Pavone AI, Sawicki J, Fischer SM. Progressive metaplastic and dysplastic changes in mouse pancreas induced by cyclooxygenase-2 overexpression. Neoplasia. 2008 Aug;10(8):782-96.
- Weiserová M, Ryu J. Characterization of a restriction modification system from the commensal Escherichia coli strain A0 34/86 (O83:K24:H31). BMC Microbiol. 2008 Jun 27;8:106. doi: 10.1186/1471-2180-8-106.
- Ryu J, Rowsell E. Quick identification of Type I restriction enzyme isoschizomers using newly developed pTypeI and reference plasmids. Nucleic Acids Res. 2008 Aug;36(13):e81. doi: 10.1093/nar/gkn056. Epub 2008 Jun 18.
- Schuhmacher AJ, Guerra C, Sauzeau V, Cañamero M, Bustelo XR, Barbacid M. A mouse model for Costello syndrome reveals an Ang II-mediated hypertensive condition. J Clin Invest. 2008 Jun;118(6):2169-79. doi: 10.1172/JCI34385.
- Hua G, Zhang R, Abdullah MA, Adang MJ. Anopheles gambiae cadherin AgCad1 binds the Cry4Ba toxin of Bacillus thuringiensis israelensis and a fragment of AgCad1 synergizes toxicity. Biochemistry. 2008 May 6;47(18):5101-10. doi: 10.1021/bi7023578. Epub 2008 Apr 12.
- Kasarjian JK, Kodama Y, Iida M, Matsuda K, Ryu J. Four new type I restriction enzymes identified in Escherichia coli clinical isolates. Nucleic Acids Res. 2005 Jul 21;33(13):e114.
- Strand SS, Leib DA. Role of the VP16-binding domain of vhs in viral growth, host shutoff activity, and pathogenesis. J Virol. 2004 Dec;78(24):13562-72.
- Kasarjian JK, Hidaka M, Horiuchi T, Iida M, Ryu J. The recognition and modification sites for the bacterial type I restriction systems KpnAI, StySEAI, StySENI and StySGI. Nucleic Acids Res. 2004 Jun 15;32(10):e82.
- Kirby AE, King ND, Connell TD. RhuR, an extracytoplasmic function sigma factor activator, is essential for heme-dependent expression of the outer membrane heme and hemoprotein receptor of Bordetella avium. Infect Immun. 2004 Feb;72(2):896-907.
- Pavlova IV, Virgin HW, Speck SH. Disruption of gammaherpesvirus 68 gene 50 demonstrates that Rta is essential for virus replication. J Virol. 2003 May;77(10):5731-9.
- Kasarjian JK, Iida M, Ryu J. New restriction enzymes discovered from Escherichia coli clinical strains using a plasmid transformation method. Nucleic Acids Res. 2003 Mar 1;31(5):e22.
- Ding L, Derdowski A, Wang JJ, Spearman P. Independent segregation of human immunodeficiency virus type 1 Gag protein complexes and lipid rafts. J Virol. 2003 Feb;77(3):1916-26.
- Murphy ER, Sacco RE, Dickenson A, Metzger DJ, Hu Y, Orndorff PE, Connell TD. BhuR, a virulence-associated outer membrane protein of Bordetella avium, is required for the acquisition of iron from heme and hemoproteins. Infect Immun. 2002 Oct;70(10):5390-403.
- Claes WA, Pühler A, Kalinowski J. Identification of two prpDBC gene clusters in Corynebacterium glutamicum and their involvement in propionate degradation via the 2-methylcitrate cycle. J Bacteriol. 2002 May;184(10):2728-39.
- François A, Eterradossi N, Delmas B, Payet V, Langlois P. Construction of avian adenovirus CELO recombinants in cosmids. J Virol. 2001 Jun;75(11):5288-301.
pMeca Plasmid utilization
If you publish research with this product, please let us know so we can cite your paper.