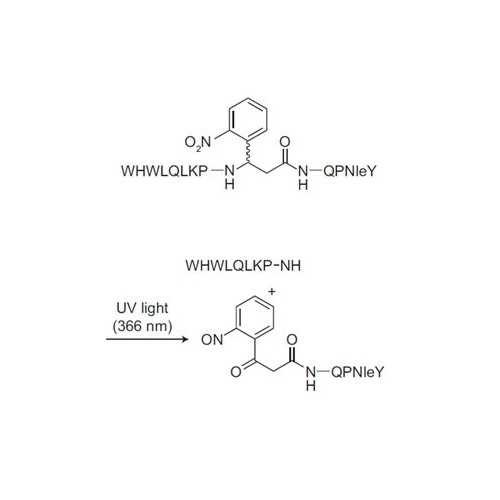
Photocleavable Alpha-Factor for Yeast
This photocleavable alpha-factor analog incorporates a UV-cleavable linker into the backbone, which enables photolysis of the peptide upon exposure to UV light. This mimics Bar1 protease cleavage and cell cycle release.
Highlights:
- Photocleavable 2-(2-nitrophenyl)-aminopropionyl group. No washing required.
- Cleavage via UV light (365 nm via handheld or chamber-based UV source, or through UV excitation filter in a microscope).
- Norleucine residue at position 12 in place of the native methionine, eliminating the potential for oxidative damage to the peptide.
- Ideal for studies where timing and synchronicity are crucial for reinitiating the yeast cell cycle, and/or when washout is not possible (e.g. microscopy-based experiments on solid media).
 Suggested Protocol for Photocleavable Alpha-Factor Arrest
Suggested Protocol for Photocleavable Alpha-Factor Arrest
The yeast pheromone peptide alpha-factor (WHWLQLKPGQPMY) binds to the G-protein coupled receptor, Ste2p. Haploid MAT-alpha yeast cells respond to this pheromone by arresting their cell cycles in the G1 phase and preparing for mating with MAT-alpha cells in the vicinity by extending cell wall projections and inducing mating-specific genes. Under native conditions, a protease called Bar1 degrades alpha-factor by cleaving at the lysine residue in order to stop the mating response. Because of its application as a tool to arrest yeast in G1, alpha-factor peptide is widely utilized—however, to release cells from arrest and allow the cell cycle to continue requires multi-step washing to remove the peptide.
From the laboratory of Laurie L. Parker, PhD, Purdue University, developed during postdoctoral research in the groups of Stephen Kron and Stephen Kent at the University of Chicago.
Read Dr. Parker's blog post, Development of Photocleavable Alpha-Factor for Yeast »
 Part of The Investigator's Annexe program.
Part of The Investigator's Annexe program.
This photocleavable alpha-factor analog incorporates a UV-cleavable linker into the backbone, which enables photolysis of the peptide upon exposure to UV light. This mimics Bar1 protease cleavage and cell cycle release.
Highlights:
- Photocleavable 2-(2-nitrophenyl)-aminopropionyl group. No washing required.
- Cleavage via UV light (365 nm via handheld or chamber-based UV source, or through UV excitation filter in a microscope).
- Norleucine residue at position 12 in place of the native methionine, eliminating the potential for oxidative damage to the peptide.
- Ideal for studies where timing and synchronicity are crucial for reinitiating the yeast cell cycle, and/or when washout is not possible (e.g. microscopy-based experiments on solid media).
![]() Suggested Protocol for Photocleavable Alpha-Factor Arrest
Suggested Protocol for Photocleavable Alpha-Factor Arrest
The yeast pheromone peptide alpha-factor (WHWLQLKPGQPMY) binds to the G-protein coupled receptor, Ste2p. Haploid MAT-alpha yeast cells respond to this pheromone by arresting their cell cycles in the G1 phase and preparing for mating with MAT-alpha cells in the vicinity by extending cell wall projections and inducing mating-specific genes. Under native conditions, a protease called Bar1 degrades alpha-factor by cleaving at the lysine residue in order to stop the mating response. Because of its application as a tool to arrest yeast in G1, alpha-factor peptide is widely utilized—however, to release cells from arrest and allow the cell cycle to continue requires multi-step washing to remove the peptide.
From the laboratory of Laurie L. Parker, PhD, Purdue University, developed during postdoctoral research in the groups of Stephen Kron and Stephen Kent at the University of Chicago.
Read Dr. Parker's blog post, Development of Photocleavable Alpha-Factor for Yeast »
 Part of The Investigator's Annexe program.
Part of The Investigator's Annexe program.
| Product Type: | Protein |
| Name: | Alpha-Factor |
| Source: | Synthetic |
| Molecular Weight: | MW = 1799.96 g/mol, FW (TFA salt) = 2096.96 |
| Amino Acid Sequence: | WHWLQLKP-(2-(2-nitrophenyl)-aminopropionyl)-QP(Nle)Y |
| Purity: | >90%. LC/MS |
| Concentration: | 1 mM in DMSO |
| Comments: | Recommended amount: 1uM (Bar1 deletion) to 10uM (WT). See protocol for details. |
| Storage: | -80C to -20C. Light sensitive, store in dark place. |
| Shipped: | Cold packs |
![]() Suggested Protocol for Photocleavable Alpha-Factor Arrest
Suggested Protocol for Photocleavable Alpha-Factor Arrest
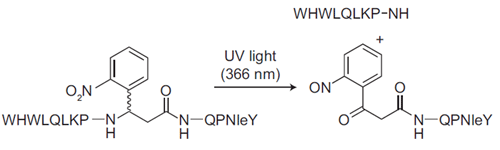
Adapted from: Parker LL, Kurutz JW, Kent SBH, and Kron SJ. Control of the Yeast Cell Cycle with a Photocleavable Alpha-Factor Analogue. Angew. Chem., Int. Ed. 2006, 38, 63226325.
Flow cytometry analysis of the release from G1 arrest by a G1/G2 trap assay.
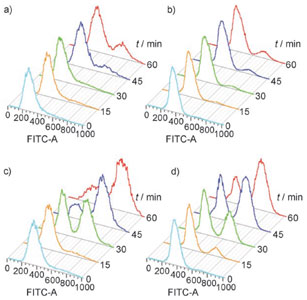
The DNA content was determined as 1N (G1 trapped) or 2N (G2 trapped; SYTOX green staining). a) Non-cleavable alpha factor (WHWLQLKPGQPNleY), no release; b) Photocleavable Alpha Factor, no release; c) Non-cleavable alpha factor, washout; d) Photocleavable Alpha Factor, UV (5 min, 366 nm, 20 mWcm^-2). FTIC-A=absorption of SYTOX green fluorescein-based DNA stain).
Adapted from: Parker LL, Kurutz JW, Kent SBH, and Kron SJ. Control of the Yeast Cell Cycle with a Photocleavable Alpha-Factor Analogue. Angew. Chem., Int. Ed. 2006, 38, 63226325.
Each 200uL unit contains enough alpha-factor for 8+ experiments in WT strains under conditions that use the highest recommended concentration in the recommended volume, 2.5 mL (more for smaller scale or Bar1-deletion experiments). See suggested protocol for more information.
- Parker LL, Kurutz JW, Kent SBH, and Kron SJ. Control of the Yeast Cell Cycle with a Photocleavable Alpha-Factor Analogue. Angew. Chem., Int. Ed. 2006, 38, 63226325.
If you publish research with this product, please let us know so we can cite your paper.