Trypanosoma cruzi CRISPR/Cas9 System
Trypanosoma cruzi CRISPR/Cas9 System plasmids allow for the amplification of a specific sgRNA sequence or express cas9 to generate CRISPR-ablated, red/green fluorescent parasites.
Highlights:
- CAS9/pTREX-n: S. pyogenes Cas9 sequence with a twice-repeated sequence of the simian virus 40 nuclear localization signal and GFP in the pTREX-n backbone. Used for cloning a specific sgRNA by BamHI, to be co-expressed with Cas9 for genome editing in Trypanosoma cruzi
- pUC_sgRNA: Contains the empty sgRNA backbone sequence (tracrRNA, 82 bp) and is used as DNA template to amplify a specific sgRNA using a forward primer with the protospacer sequence for gene targeting
- td Tomato/pTREX-b: Possesses tdTomato synthetic gene and blasticidin S deaminase (Bsd) gene and can be used for cloning a specific sgRNA by BamHI, and then transfect Trypanosoma cruzi epimastigotes expressing Cas9 (green fluorescent and neomycin resistant) to generate CRISPR-ablated, red/green fluorescent parasites
From the laboratory of Roberto Docampo, MD, PhD, University of Georgia.
 Part of The Investigator's Annexe program.
Part of The Investigator's Annexe program.
Trypanosoma cruzi CRISPR/Cas9 System plasmids allow for the amplification of a specific sgRNA sequence or express cas9 to generate CRISPR-ablated, red/green fluorescent parasites.
Highlights:
- CAS9/pTREX-n: S. pyogenes Cas9 sequence with a twice-repeated sequence of the simian virus 40 nuclear localization signal and GFP in the pTREX-n backbone. Used for cloning a specific sgRNA by BamHI, to be co-expressed with Cas9 for genome editing in Trypanosoma cruzi
- pUC_sgRNA: Contains the empty sgRNA backbone sequence (tracrRNA, 82 bp) and is used as DNA template to amplify a specific sgRNA using a forward primer with the protospacer sequence for gene targeting
- td Tomato/pTREX-b: Possesses tdTomato synthetic gene and blasticidin S deaminase (Bsd) gene and can be used for cloning a specific sgRNA by BamHI, and then transfect Trypanosoma cruzi epimastigotes expressing Cas9 (green fluorescent and neomycin resistant) to generate CRISPR-ablated, red/green fluorescent parasites
From the laboratory of Roberto Docampo, MD, PhD, University of Georgia.
 Part of The Investigator's Annexe program.
Part of The Investigator's Annexe program.
| Catalog Number | Product | DataSheet | Size | AVAILABILITY | Price | Qty |
|---|
| Product Type: | Plasmid |
| Name: | CAS9/pTREX-n |
| Gene/insert name: | Fusion gene CAS9-HA-2xNLS-GFP |
| Antibiotic Resistance: | Ampicillin |
| Fusion Tag(s): | Fusion gene Cas9-HA-2xNLS-GFP |
| Grow in E. coli at 37 C: | DH5alpha; 37C |
| Selectable markers: | Neomycin (select with G418) |
| Cloning Site 5': | XbaI |
| Cloning Site 3': | HindIII |
| Insert Size: | 4975 bp |
| Vector Backbone and Size: | pTREX-n, backbone size without insert: 6227bp |
| High or low copy: | High |
| Storage: | -20C |
| Shipped: | Ambient temperature |
Trypanosoma cruzi CRISPR/Cas9 System Plasmids
| Plasmid: | CAS9/pTREX-n | PUC_sgRNA | tdTomato/pTREX-b |
| Gene/Insert Name: | Fusion gene CAS9-HA-2xNLS-GFP | TracrRNA sequence | tdTomato |
| Insert Size (bp): | 4975 | 82 | 1430 |
| Species: | Synthetic; Streptococcus pyogenes | N/A | Synthetic |
| Fusion Proteins/Tags: | Fusion gene Cas9-HA-2xNLS-GFP | N/A | |
| Vector Backbone and Size: | pTREX-n, backbone size without insert: 6227bp | pUCAmp; 3150bp | pTREX-b, backbone size without insert: ~5868bp |
| Cloning Site 5': | XbaI | 5' Sequencing Primer: M13_fwd20_primer and M13_pUC_fwd_primer | XbaI |
| Cloning Site 3': | HindIII | 3' Sequencing Primer: M13_rev_primer and M13_pUC_rev_primer | HindIII |
| Antibiotic Resistance: | Ampicillin | Ampicillin | Ampicillin |
| High or Low Copy: | High | High | High |
| Grow in Standard E. coli @ 37C: | DH5alpha; 37C | DH5alpha; 37C | DH5alpha; 37C |
| Selectable Markers: | Neomycin (select with G418) | Blasticidin | |
| Storage Temperature: | -20C | -20C | -20C |
| Shipped: | Room Temperature | Room Temperature | Room Temperature |
CAS9/pTREX-n
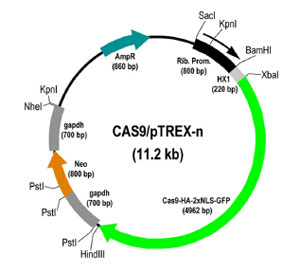
Plasmid map for CAS9-HA-2xNLS-GFP in pTREX-n. Restriction map of molecular constructs generated for the CRISPR/Cas9 system in T. cruzi. Cas9/pTREX-n derived from vectors pTREX-n by insertion of the Cas9-HA-2 NLS-GFP fusion gene through XbaI and HindIII restriction sites.
tdTomato/pTREX-b
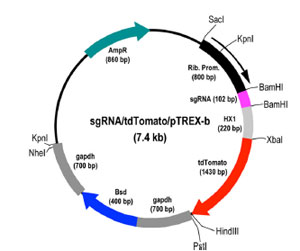
Plasmid map for td Tomato in pTREX-b_NL. Plasmid contains cytoplasmic red fluorescence and blasticidin resistance and can be used to transfect T. cruzi epimastigotes expressing Cas9 (green fluorescent and neomycin resistant) to generated CRISPR-ablated, red/green fluorescent parasites.
Adapted from: Lander N. et al., mBIO. 2015. 6(4): e01012-15.
- Lander N, Li ZH, Niyogi S, Docampo R. CRISPR/Cas9-Induced Disruption of Paraflagellar Rod Protein 1 and 2 Genes in Trypanosoma cruzi Reveals Their Role in Flagellar Attachment. mBIO. 2015. 6(4): e01012-15.
If you publish research with this product, please let us know so we can cite your paper.


