SpyTag/SpyCatcher Protein Coupling Reagents
The SpyTag/SpyCatcher system is a convenient protein coupling tool for irreversible peptide-protein ligation. It is ideal for binding, labeling, immobilization and creating new kinds of protein architectures.
Highlights:
- SpyTag forms a spontaneous amide bond upon binding its genetically encoded partner SpyCatcher
- Highly compatible - SpyTag reacts with SpyCatcher under a wide range of conditions
- Extremely stable reaction product - SpyCatcher/SpyTag complex is stable to boiling in SDS
- SpyCatcher (S49C) version contains a unique cysteine residue for precise labeling with dye or precise attachment to surfaces or beads
Peptide interaction with proteins is usually weak. SpyTag is a genetically encoded peptide (SpyTag003 sequence RGVPHIVMVDAYKRYK) that forms a spontaneous amide bond upon binding its genetically encoded partner SpyCatcher (with the latest version SpyCatcher003 available here). SpyTag003 reacts with SpyCatcher003 under a wide range of conditions and the after reaction the product is stable to boiling in SDS. SpyCatcher003 (S49C) has a unique cysteine for precise labeling with dye or precise attachment to surfaces or beads.
From the laboratory of Mark Howarth, PhD, University of Oxford.
The SpyTag/SpyCatcher system is a convenient protein coupling tool for irreversible peptide-protein ligation. It is ideal for binding, labeling, immobilization and creating new kinds of protein architectures.
Highlights:
- SpyTag forms a spontaneous amide bond upon binding its genetically encoded partner SpyCatcher
- Highly compatible - SpyTag reacts with SpyCatcher under a wide range of conditions
- Extremely stable reaction product - SpyCatcher/SpyTag complex is stable to boiling in SDS
- SpyCatcher (S49C) version contains a unique cysteine residue for precise labeling with dye or precise attachment to surfaces or beads
Peptide interaction with proteins is usually weak. SpyTag is a genetically encoded peptide (SpyTag003 sequence RGVPHIVMVDAYKRYK) that forms a spontaneous amide bond upon binding its genetically encoded partner SpyCatcher (with the latest version SpyCatcher003 available here). SpyTag003 reacts with SpyCatcher003 under a wide range of conditions and the after reaction the product is stable to boiling in SDS. SpyCatcher003 (S49C) has a unique cysteine for precise labeling with dye or precise attachment to surfaces or beads.
From the laboratory of Mark Howarth, PhD, University of Oxford.
| Catalog Number | Product | DataSheet | Size | AVAILABILITY | Price | Qty |
|---|
| Product Type: | Protein |
| Name: | SpyTag/SpyCatcher Protein Coupling Reagents |
| Accession ID: | JQ478411.1 (SpyCatcher) |
| Source: | Recombinant expression in E. coli |
| Molecular Weight: | 15,597.7 Da (SpyCatcher003) 15,613.7 Da (SpyCatcher003, S49C); 44,867.78 Da (SpyTag003-MBP) |
| Amino Acid Sequence: |
SpyCatcher003: SYYHHHHHHDYDIPTTENLYFQGAMVTTLSGLSGEQGPSGDMTTEEDSATHIKFSKRDEDGRELAGATMELRDSSGKTISTWISDGHVKDFYLYPGKYTFVETAAPDGYEVATPIEFTVNEDGQVTVDGEATEGDAHTGSSGS SpyCatcher003 (S49C): SYYHHHHHHDYDIPTTENLYFQGAMVTTLSGLSGEQGPSGDMTTEEDSATHIKFSKRDEDGRELAGATMELRDCSGKTISTWISDGHVKDFYLYPGKYTFVETAAPDGYEVATPIEFTVNEDGQVTVDGEATEGDAHTGSSGS SpyTag003: GSSHHHHHHSSGLVPRGSRGVPHIVMVDAYKRYKGSGESGKIEEGKLVIWINGDKGYNGLAEVGKKFEKDTGIKVTVEHPDKLEEKFPQVAATGDGPDIIFWAHDRFGGYAQSGLLAEITPDKAFQDKLYPFTWDAVRYNGKLIAYPIAVEALSLIYNKDLLPNPPKTWEEIPALDKELKAKGKSALMFNLQEPYFTWPLIAADGGYAFKYENGKYDIKDVGVDNAGAKAGLTFLVDLIKNKHMNADTDYSIAEAAFNKGETAMTINGPWAWSNIDTSKVNYGVTVLPTFKGQPSKPFVGVLSAGINAASPNKELAKEFLENYLLTDEGLEAVNKDKPLGAVALKSYEEELAKDPRIAATMENAQKGEIMPNIPQMSAFWYAVRTAVINAASGRQTVDEALKDAQTNSSS |
| Fusion Tag(s): | N-terminal 6xHis tag for each |
| Purity: | >95%. Purified from Ni-NTA affinity chromatography and then by size exclusion chromatography. |
| Buffer: | Phosphate buffered saline (PBS) pH 7.4 |
| Concentration: | 2.32mg/mL (SpyCatcher003), 1.8mg/mL (SpyCatcher003, S49C), 2.47mg/mL (SpyTag003-MBP) |
| Amount: | 0.5mg |
| Storage: | -80C, avoid multiple freeze-thaw cycles |
| Shipped: | Dry ice |
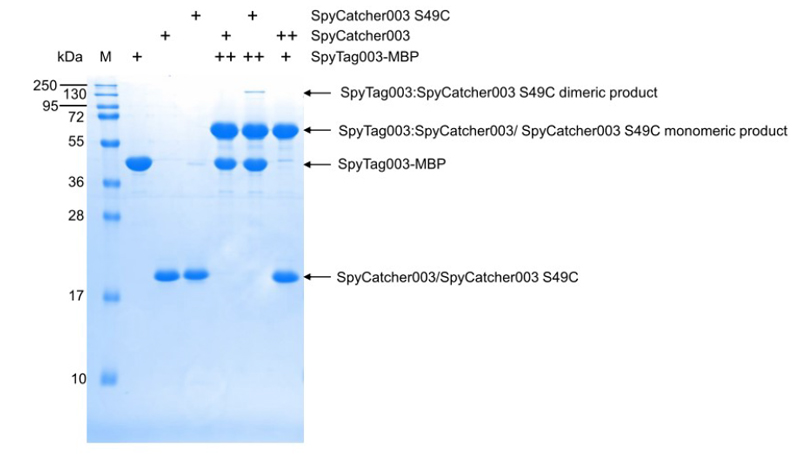
SpyCatcher003 reacts with SpyTag in the pH range 5-8 and is relatively insensitive to the buffer composition, as long as SDS or denaturants like urea or guanidinium are not used.
SpyCatcher003: SDS-PAGE shows one higher band from gluconylation, which does not affect reactivity with SpyTag.
- Theoretical pI: 4.52
- Extinction coefficient at 280 nm (predicted by ProtParam): 17,420 M-1 cm-1
SpyCatcher003 (S49C): SDS-PAGE shows a 2nd band at slightly higher MW related to glucosylation, but this does not affect reactivity. As expected for exposed cysteines, SpyCatcher003 (S49C) will eventually form a disulfide upon storage. This disulfide-bonded form will not be reactive with maleimide or other thiol-reactive reagents. To test the extent of disulfide bond formation, perform SDS-PAGE with Coomassie staining with and without DTT.
- Theoretical pI: 4.52
- Extinction coefficient at 280 nm (predicted by ProtParam): 17,420 M-1 cm-1
SpyTag003-MBP:
- Theoretical pI: 5.87
- Extinction coefficient at 280 nm (predicted by ProtParam): 69,330 M-1 cm-1
- Keeble AH, Turkki P, Stokes S, Khairil Anuar INA, Rahikainen R, Hytönen VP, Howarth M. Approaching infinite affinity through engineering of peptide-protein interaction. Proc Natl Acad Sci U S A. 2019 Dec 10;116(52):26523-26533.
- Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, Howarth M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci U S A. 2012 Mar 20;109(12):E690-7.
- Veggiani G., Zakeri B., Howarth M. Superglue from Bacteria: Unbreakable Bridges for Protein Nanotechnology. Trends in Biotechnology 2014 Oct;32(10):506-12.
- Schoene C, Fierer JO, Bennett SP, Howarth M. SpyTag/SpyCatcher Cyclization Confers Resilience to Boiling on a Mesophilic Enzyme. Angewandte Chemie. 2014 Jun 10;53(24):6101-4.
- Fierer JO, Veggiani G, Howarth M. SpyLigase peptide-peptide ligation polymerizes affibodies to enhance magnetic cancer cell capture. Proc Natl Acad Sci U S A. 2014 Apr 1;111(13):E1176-81.
- Li L, Fierer JO, Rapoport TA, Howarth M. Structural analysis and optimization of the covalent association between SpyCatcher and a peptide tag. Journal of Molecular Biology. 2014 Jan 23;426(2):309-17.
- Min, Duyoung; Arbing, Mark A; Jefferson, Robert E; Bowie, James U. A simple DNA handle attachment method for single molecule mechanical manipulation experiments.Protein Sci. 2016 01;25(8):1535-44 View Article
- Dovala, Dustin; Sawyer, William S; Rath, Christopher M; Metzger, Louis E. Rapid analysis of protein expression and solubility with the SpyTag-SpyCatcher system.Protein Expr Purif. 2016 01;117:44-51 View Article
- Alves, Nathan J; Turner, Kendrick B; Daniele, Michael A; Oh, Eunkeu; Medintz, Igor L; Walper, Scott A. Bacterial Nanobioreactors--Directing Enzyme Packaging into Bacterial Outer Membrane Vesicles.ACS Appl Mater Interfaces. 2015 11 11;7(44):24963-72 View Article
- Reddington, Samuel C; Howarth, Mark. Secrets of a covalent interaction for biomaterials and biotechnology: SpyTag and SpyCatcher.Curr Opin Chem Biol. 2015 12;29:94-9 View Article
- Janitzek, Christoph M; Matondo, Sungwa; Thrane, Susan; Nielsen, Morten A; Kavishe, Reginald; Mwakalinga, Steve B; Theander, Thor G; Salanti, Ali; Sander, Adam F. Bacterial superglue generates a full-length circumsporozoite protein virus-like particle vaccine capable of inducing high and durable antibody responses.Malar J. 2016 11 08;15(1):545 View Article
- Si, Meng; Xu, Qing; Jiang, Ling; Huang, He. SpyTag/SpyCatcher Cyclization Enhances the Thermostability of Firefly Luciferase.PLoS One. 2016;11(9):e0162318 View Article
- Zhang, Wen-Bin; Sun, Fei; Tirrell, David A; Arnold, Frances H. Controlling macromolecular topology with genetically encoded SpyTag-SpyCatcher chemistry.J Am Chem Soc. 2013 09 18;135(37):13988-97 View Article
- Fairhead, Michael; Veggiani, Gianluca; Lever, Melissa; Yan, Jun; Mesner, Dejan; Robinson, Carol V; Dushek, Omer; Van der Merwe, P Anton; Howarth, Mark. SpyAvidin hubs enable precise and ultrastable orthogonal nanoassembly.J Am Chem Soc. 2014 09 03;136(35):12355-63 View Article
- Tan, Lee Ling; Hoon, Shawn S; Wong, Fong T. Kinetic Controlled Tag-Catcher Interactions for Directed Covalent Protein Assembly.PLoS One. 2016;11(10):e0165074 View Article
- Wang, Jindan; Wang, Yilin; Wang, Xinzhe; Zhang, Dandan; Wu, Shuyu; Zhang, Guangya. Enhanced thermal stability of lichenase from Bacillus subtilis 168 by SpyTag/SpyCatcher-mediated spontaneous cyclization.Biotechnol Biofuels. 2016;9:79 View Article
- Thrane, Susan; Janitzek, Christoph M; Matondo, Sungwa; Resende, Mafalda; Gustavsson, Tobias; De Jongh, Willem Adriaan; Clemmensen, Stine; Roeffen, Will; Van de Vegte-Bolmer, Marga; Van Gemert, Geert Jan; Sauerwein, Robert; Schiller, John T; Nielsen, Morten A; Theander, Thor G; Salanti, Ali; Sander, Adam F. Bacterial superglue enables easy development of efficient virus-like particle based vaccines.J Nanobiotechnology. 2016 04 27;14:30 View Article
- Bedbrook, Claire N; Kato, Mihoko; Ravindra Kumar, Sripriya; Lakshmanan, Anupama; Nath, Ravi D; Sun, Fei; Sternberg, Paul W; Arnold, Frances H; Gradinaru, Viviana. Genetically Encoded Spy Peptide Fusion System to Detect Plasma Membrane-Localized Proteins In Vivo.Chem Biol. 2015 08 20;22(8):1108-21 View Article
- Sun, Fei; Zhang, Wen-Bin; Mahdavi, Alborz; Arnold, Frances H; Tirrell, David A. Synthesis of bioactive protein hydrogels by genetically encoded SpyTag-SpyCatcher chemistry.Proc Natl Acad Sci U S A. 2014 08 05;111(31):11269-74 View Article
- Brune, Karl D; Leneghan, Darren B; Brian, Iona J; Ishizuka, Andrew S; Bachmann, Martin F; Draper, Simon J; Biswas, Sumi; Howarth, Mark. Plug-and-Display: decoration of Virus-Like Particles via isopeptide bonds for modular immunization.Sci Rep. 2016 01 19;6:19234 View Article
- Veggiani, Gianluca; Nakamura, Tomohiko; Brenner, Michael D; Gayet, Raphaël V; Yan, Jun; Robinson, Carol V; Howarth, Mark. Programmable polyproteams built using twin peptide superglues.Proc Natl Acad Sci U S A. 2016 02 02;113(5):1202-7 View Article
- Schoene, Christopher; Bennett, S Paul; Howarth, Mark. SpyRing interrogation: analyzing how enzyme resilience can be achieved with phytase and distinct cyclization chemistries.Sci Rep. 2016 02 10;6:21151 View Article
- Brune, Karl; Buldun, Can; Li, Yuanyuan; Taylor, Iona; Brod, Florian; Biswas, Sumi; Howarth, Mark. Dual plug-and-display synthetic assembly using orthogonal reactive proteins for twin antigen immunization.Bioconjug Chem. 2017 04 24; View Article
- Gao, Xiaoye; Fang, Jie; Xue, Bin; Fu, Linglan; Li, Hongbin. Engineering Protein Hydrogels Using SpyCatcher-SpyTag Chemistry.Biomacromolecules. 2016 09 12;17(9):2812-9 View Article
- Schoene, C; Bennett, S P; Howarth, M. SpyRings Declassified: A Blueprint for Using Isopeptide-Mediated Cyclization to Enhance Enzyme Thermal Resilience.Methods Enzymol. 2016;580:149-67 View Article
- Botyanszki, Zsofia; Tay, Pei Kun R; Nguyen, Peter Q; Nussbaumer, Martin G; Joshi, Neel S. Engineered catalytic biofilms: Site-specific enzyme immobilization onto E. coli curli nanofibers.Biotechnol Bioeng. 2015 10;112(10):2016-24 View Article
- Liu, Zhida; Zhou, Hang; Wang, Wenjun; Tan, Wenjie; Fu, Yang-Xin; Zhu, Mingzhao. A novel method for synthetic vaccine construction based on protein assembly.Sci Rep. 2014 12 01;4:7266 View Article
- Pröschel, Marlene; Detsch, Rainer; Boccaccini, Aldo R; Sonnewald, Uwe. Engineering of Metabolic Pathways by Artificial Enzyme Channels.Front Bioeng Biotechnol. 2015;3:168 View Article
- Siegmund, Vanessa; Piater, Birgit; Zakeri, Bijan; Eichhorn, Thomas; Fischer, Frank; Deutsch, Carl; Becker, Stefan; Toleikis, Lars; Hock, Björn; Betz, Ulrich a K; Kolmar, Harald. Spontaneous Isopeptide Bond Formation as a Powerful Tool for Engineering Site-Specific Antibody-Drug Conjugates.Sci Rep. 2016 12 16;6:39291 View Article
- Wang, Hejia Henry; Altun, Burcin; Nwe, Kido; Tsourkas, Andrew. Proximity-Based Sortase-Mediated Ligation.Angew Chem Int Ed Engl. 2017 05 02;56(19):5349-5352 View Article
- Alves, Nathan J; Turner, Kendrick B; Medintz, Igor L; Walper, Scott A. Protecting enzymatic function through directed packaging into bacterial outer membrane vesicles.Sci Rep. 2016 04 27;6:24866 View Article
- Chen, Allen Y; Deng, Zhengtao; Billings, Amanda N; Seker, Urartu O S; Lu, Michelle Y; Citorik, Robert J; Zakeri, Bijan; Lu, Timothy K. Synthesis and patterning of tunable multiscale materials with engineered cells.Nat Mater. 2014 05;13(5):515-23 View Article
- Regan, Lynne; Caballero, Diego; Hinrichsen, Michael R; Virrueta, Alejandro; Williams, Danielle M; O'hern, Corey S. Protein design: Past, present, and future.Biopolymers. 2015 07;104(4):334-50 View Article
- Leonard, John D; Narlikar, Geeta J. A nucleotide-driven switch regulates flanking DNA length sensing by a dimeric chromatin remodeler.Mol Cell. 2015 03 05;57(5):850-9 View Article
- Liu, Xueliang; Lopez, Paola A; Giessen, Tobias W; Giles, Michael; Way, Jeffrey C; Silver, Pamela A. Engineering Genetically-Encoded Mineralization and Magnetism via Directed Evolution.Sci Rep. 2016 11 29;6:38019 View Article
- Schmid-Burgk, Jonathan L; Höning, Klara; Ebert, Thomas S; Hornung, Veit. CRISPaint allows modular base-specific gene tagging using a ligase-4-dependent mechanism.Nat Commun. 2016 07 28;7:12338 View Article
- Zakeri, Bijan; Lu, Timothy K. Synthetic biology of antimicrobial discovery.ACS Synth Biol. 2013 07 19;2(7):358-72 View Article
- Alam MK, El-Sayed A, Barreto K, Bernhard W, Fonge H, Geyer CR. Site-Specific Fluorescent Labeling of Antibodies and Diabodies Using SpyTag/SpyCatcher System for In Vivo Optical Imaging. Mol Imaging Biol. 2018 Jun 12. View Article
If you publish research with this product, please let us know so we can cite your paper.


