Biotinylated sRNA Ladder
Highly sensitive biotinylated RNA standard suitable for bacterial small RNA (sRNA) research.
Highlights:
- Easily determine the size of bacterial sRNA without radiation
- Highly sensitive probe (less than or ca. 12 ng/µL)
- Suitable for sRNA bacterial and Archaea research and Northern Blot analysis
The discovery of bacterial small RNA (sRNA) is a major area of research in microbiology. Northern blotting remains the gold standard for sRNA identification since it confirms presence and size. To determine size using a northern blot it is necessary to run an RNA standard of known size. Typically, estimating the size of sRNA requires adding radioactive [U-32P]ATP to an RNA ladder using T4 polynucleotide kinase and imaging the resulting blot with film or a radioimager. The Biotinylated sRNA Ladder allows for a non-radioactive method of northern blotting using biotin linked probes and RNA ladders to identify and size small RNAs. The biotin linked probes work by adding a streptavidin-horsradish peroxidase conjugate to the blot then adding luminol and hydrogen peroxide to create chemiluminescence.
From the laboratory of Mary Ann Moran, PhD, Andrew S. Burns, PhD and Adam Rivers, PhD, University of Georgia.
 Part of The Investigator's Annexe program.
Part of The Investigator's Annexe program.
Highly sensitive biotinylated RNA standard suitable for bacterial small RNA (sRNA) research.
Highlights:
- Easily determine the size of bacterial sRNA without radiation
- Highly sensitive probe (less than or ca. 12 ng/µL)
- Suitable for sRNA bacterial and Archaea research and Northern Blot analysis
The discovery of bacterial small RNA (sRNA) is a major area of research in microbiology. Northern blotting remains the gold standard for sRNA identification since it confirms presence and size. To determine size using a northern blot it is necessary to run an RNA standard of known size. Typically, estimating the size of sRNA requires adding radioactive [U-32P]ATP to an RNA ladder using T4 polynucleotide kinase and imaging the resulting blot with film or a radioimager. The Biotinylated sRNA Ladder allows for a non-radioactive method of northern blotting using biotin linked probes and RNA ladders to identify and size small RNAs. The biotin linked probes work by adding a streptavidin-horsradish peroxidase conjugate to the blot then adding luminol and hydrogen peroxide to create chemiluminescence.
From the laboratory of Mary Ann Moran, PhD, Andrew S. Burns, PhD and Adam Rivers, PhD, University of Georgia.
 Part of The Investigator's Annexe program.
Part of The Investigator's Annexe program.
| Product Type: | Buffer or Chemical |
| Name: | Biotinylated sRNA ladder |
| Buffer: | Liquid in TE buffer |
| Marker Size: | 100, 200, 300, 400, 500, 750, 1000 bases |
| Concentration: | 12 ng/uL |
| Comments: | For running of polyacrylamide gels, the Biotinylated sRNA Ladder (12ng/uL) should be mixed with equal volume of a RNA loading buffer. Then, use 1 µl of of the resulting solution per lane. |
| Storage: | -80C |
| Shipped: | Dry ice |
Absorbance of sRNA Ladder
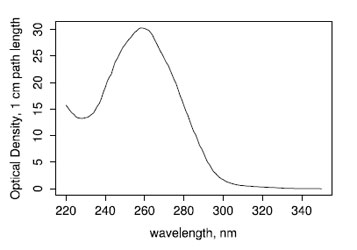
The purity and concentration of the standard was deteremined using a Nanodrop ND-1000. The volume of the stock is 100 uL the RNA is estimated at 1200 ng/uL with a 260nm/280nm ratio of 1.93 and a 260nm/230nm ratio of 2.26. The total amount of RNA was 120 ug.
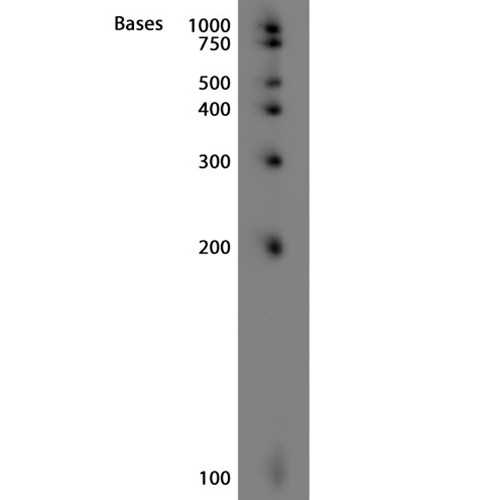
Northern Blot
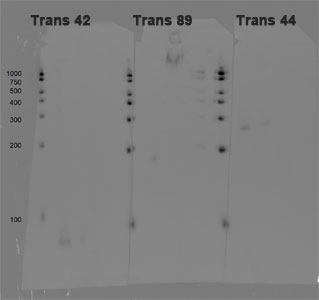
Northern blots of small RNAs from Rugerio Pomeroyi DSS-3. 1uL of 12 ng/uL standard or 20 ug of total bacterial RNA was added to each lane.
For running of polyacrylamide gels, the Biotinylated sRNA Ladder (12ng/uL) should be mixed with equal volume of a RNA loading buffer. Then, use 1 µl of of the resulting solution per lane.
- Burns AS, Bullock HA, Smith C, Huang Q, Whitman WB, Moran MA. Small RNAsexpressed during dimethylsulfoniopropionate degradation by a model marinebacterium. Environ Microbiol Rep. 2016 Jun 23. View Article
- Stacey SD, Pritchett CL. Pseudomonas aeruginosa AlgU Contributes toPosttranscriptional Activity by Increasing rsmA Expression in a mucA22 Strain. J Bacteriol. 2016 Jun 13;198(13):1812-26. View Article
- Rivers, A. R., Burns, A. S., Chan, L.-K., and Moran, M. A. (2016). Experimental Identification of Small Non-Coding RNAs in the Model Marine Bacterium Ruegeria pomeroyi DSS-3. Front. Microbiol. 7. doi:10.3389/fmicb.2016.00380.
- Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011 Dec 1;3(12).
- Locati MD, Pagano JF, Ensink WA, van Olst M, van Leeuwen S, Nehrdich U, Zhu K, Spaink HP, Girard G, Rauwerda H, Jonker MJ, Dekker RJ, Breit TM. Linking Maternal and Somatic 5S rRNA types with Different Sequence-Specific Non-LTR Retrotransposons. RNA. 2016 Dec 21. pii: rna.059642.116. View Article
- Stacey SD, Williams DA, Pritchett CL. The Pseudomonas aeruginosa two-component regulator AlgR directly activates rsmA expression in a phosphorylation independent manner. J Bacteriol. 2017 Mar 20. pii: JB.00048-17. View Article
- Locati MD, Pagano JFB, Girard G, Ensink WA, van Olst M, van Leeuwen S, Nehrdich U, Spaink HP, Rauwerda H, Jonker MJ, Dekker RJ, Breit TM. Expression of distinct maternal and somatic 5.8S, 18S, and 28S rRNA types during zebrafish development. RNA. 2017 Aug;23(8):1188-1199. View Article
- Pan L, Gardner CL, Pagliai FA, Gonzalez CF, Lorca GL. Identification of the Tolfenamic Acid Binding Pocket in PrbP from Liberibacter asiaticus. Front Microbiol. 2017 Aug 23;8:1591. View Article
- Pan L, da Silva D, Pagliai FA, Harrison NA, Gonzalez CF, Lorca GL. The Ferredoxin-Like Protein FerR Regulates PrbP Activity in Liberibacter asiaticus. Appl Environ Microbiol. 2019 Feb 6;85(4). pii: e02605-18. doi: 10.1128/AEM.02605-18. View Article
If you publish research with this product, please let us know so we can cite your paper.