Cytoplasmic [NiFe]-Hydrogenase (SHI)
This recombinant Pyrococcus furiosus NADP-specific NiFe-type cytoplasmic hydrogenase (SHI) is from the hyperthermophilic P. furiosus microorganism that grows optimally at 100°C. The enzyme is active over a broad temperature range, from ambient to 100°C, and is oxygen-sensitive, with a half-life in air of approximately 25 hours at room temperature.
Highlights:
- Purity of >90%
- Highly thermostable - active over broad range (up to 100°C)
- Advantageous over other enzymatic methods to regenerate reduced co-factors, as they don't generate oxidation co-products other than H+
The [NiFe] hydrogenases contain a minimum of two subunits known as the small (S) and large (L) subunits. The small subunit contains three iron-sulfur clusters while the large subunit contains the active site, a nickel-iron center which is connected to the solvent by a molecular tunnel. To date, periplasmic, cytoplasmic, and membrane-bound hydrogenases have been found. [NiFe] hydrogenases are known to be deactivated by molecular oxygen (O2). The [NiFe] hydrogenase of Pyrococcus furiosus is heterotetrameric wherein the additional two subunits allow the enzyme to use NAD(P)(H) as an electron carrier.
From the laboratory of Michael W. W. Adams, PhD, University of Georgia.
 Part of The Investigator's Annexe program.
Part of The Investigator's Annexe program.
This recombinant Pyrococcus furiosus NADP-specific NiFe-type cytoplasmic hydrogenase (SHI) is from the hyperthermophilic P. furiosus microorganism that grows optimally at 100°C. The enzyme is active over a broad temperature range, from ambient to 100°C, and is oxygen-sensitive, with a half-life in air of approximately 25 hours at room temperature.
Highlights:
- Purity of >90%
- Highly thermostable - active over broad range (up to 100°C)
- Advantageous over other enzymatic methods to regenerate reduced co-factors, as they don't generate oxidation co-products other than H+
The [NiFe] hydrogenases contain a minimum of two subunits known as the small (S) and large (L) subunits. The small subunit contains three iron-sulfur clusters while the large subunit contains the active site, a nickel-iron center which is connected to the solvent by a molecular tunnel. To date, periplasmic, cytoplasmic, and membrane-bound hydrogenases have been found. [NiFe] hydrogenases are known to be deactivated by molecular oxygen (O2). The [NiFe] hydrogenase of Pyrococcus furiosus is heterotetrameric wherein the additional two subunits allow the enzyme to use NAD(P)(H) as an electron carrier.
From the laboratory of Michael W. W. Adams, PhD, University of Georgia.
 Part of The Investigator's Annexe program.
Part of The Investigator's Annexe program.
| Product Type: | Protein |
| Name: | Cytoplasmic [NiFe]-Hydrogenase (4 subunits; OE-SHI) |
| Accession ID: | PF0894, NP_578623.1, PF0893, NP_578622.1, PF0892, NP_578621.1, PF0891, NP_578620.1 |
| Source: | Pyrococcus furiosus |
| Molecular Weight: | Predicted: 155 kDa, Size Exclusion: 149 kDa +/- 5 kDa |
| Amino Acid Sequence: | Sequences available upon request |
| Fusion Tag(s): | His(x9): HHHHHHHHHG |
| Purity: | >90% |
| Buffer: | 50 mM Tris, 2 mM DT, 300 mM NaCl, pH 8.2 |
| Concentration: | 1mg/mL (>100 U/mL, see Additional Information below) |
| Activity: | One unit (U) is 1 umole of H2 evolved min-1 mg-1 using MV-linked assay. |
| Comments: | NADP(H)-dependent reversible hydrogenase |
| Storage: | This item is oxygen sensitive. Stable when stored sealed in strictly anaerobic environment (<10 ppm O2) at room temperature for up to 6 months. For long-term storage, protein can be flash frozen in nitrogen and stored at -80C. |
| Shipped: | Dry ice |
Additional Information
This is a Histidine-tagged version of the published Strep-tag II version. This is an extremely thermostable, but oxygen sensitive enzyme. One unit (U) is 1 µmole of H2 evolved min-1 mg-1 using MV-linked assay.
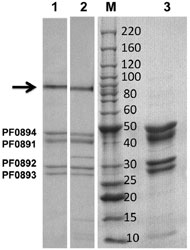
The purified enzyme was analyzed by conventional SDS-PAGE except that the protein was incubated with the SDS-loading buffer for 10 min (lane 1) or for 60 min (lane 3) prior to electrophoresis. Native SHI (treated for 10 min) is shown in lane 2. The arrow indicates the high molecular weight catalytically active protein band seen in lanes 1 and 2. The center lane (M) contains the protein molecular weight ladder (Invitrogen) with corresponding masses as indicated in kDa.
Adapted from: Chandrayan, S. K., et al. J. Biol. Chem. 287, 3257-64.
- Ma, K. and Adams, M. W. W. (2001) Hydrogenases I and II from Pyrococcus furiosus Meths. Enzymol. 331, 208-216
- Chandrayan, S. K., McTernan, P. M., Hopkins, R. C., Sun, J., Jenney, F. E., Jr. & Adams, M. W. (2012) Engineering hyperthermophilic archaeon Pyrococcus furiosus to overproduce its cytoplasmic [NiFe]-hydrogenase J. Biol. Chem. 287, 3257-64
- Hopkins, R.C., Sun, J., Jenney, F.E., Chandrayan, S.K., McTernan, P.M., and Adams, M.W.W., (2011) "Homologous expression of a subcomplex of Pyrococcus furiosus hydrogenase that interacts with pyruvate ferredoxin oxioreductase" PLoS ONE 6(1), e26569
- Patent Pending - University of Georgia research Foundation, Inc. (WO 2009075798)
If you publish research with this product, please let us know so we can cite your paper.

![Cytoplasmic [NiFe]-Hydrogenase (SHI) Cytoplasmic [NiFE]-Hyrdogenase (SHI)](https://www.kerafast.com/MediaStorage/Product/Images/Medium/837_200120200126238360.jpg)
