LOBSTR E. coli Expression Strain
LOBSTR (low background strain) is an E. coli strain for the expression of recombinant polyhistidine-tagged proteins. This strain has been optimized for one-step downstream polyhistidine-tag affinity purification and is ideal for poorly expressing proteins.
Highlights:
- Yields recombinant polyhistidine-tagged protein of higher purity by reducing the contamination by E. coli ArnA and SlyD
- Allows for a one-step purification to eliminate the major E. coli contaminants
- Based on BL21(DE3) - use the same as other commercially available competent cells
- BL21(DE3)-RIL version - Contains extra copies of the argU, ileY, and leuW tRNA genes, as well as a chloramphenicol marker
- Ideal for purification of challenging low-expressing protein targets
A major drawback of polyhistidine-tag affinity purification of proteins expressed in E. coli is the presence of naturally histidine-rich proteins, resulting in co-purification of these contaminants. In LOBSTR, ArnA and SlyD, the two most common E. coli contaminants have been modified based on surface engineering. LOBSTR maintains normal cell growth but significantly reduces the polyhistidine-tag binding affinities of ArnA and SlyD. Compared to other expression strains, LOBSTR yields recombinant protein of higher purity, allowing for one-step purifications of low expressing recombinant proteins.
From the laboratory of Thomas U. Schwartz, PhD, Massachusetts Institute of Technology.
LOBSTR (low background strain) is an E. coli strain for the expression of recombinant polyhistidine-tagged proteins. This strain has been optimized for one-step downstream polyhistidine-tag affinity purification and is ideal for poorly expressing proteins.
Highlights:
- Yields recombinant polyhistidine-tagged protein of higher purity by reducing the contamination by E. coli ArnA and SlyD
- Allows for a one-step purification to eliminate the major E. coli contaminants
- Based on BL21(DE3) - use the same as other commercially available competent cells
- BL21(DE3)-RIL version - Contains extra copies of the argU, ileY, and leuW tRNA genes, as well as a chloramphenicol marker
- Ideal for purification of challenging low-expressing protein targets
A major drawback of polyhistidine-tag affinity purification of proteins expressed in E. coli is the presence of naturally histidine-rich proteins, resulting in co-purification of these contaminants. In LOBSTR, ArnA and SlyD, the two most common E. coli contaminants have been modified based on surface engineering. LOBSTR maintains normal cell growth but significantly reduces the polyhistidine-tag binding affinities of ArnA and SlyD. Compared to other expression strains, LOBSTR yields recombinant protein of higher purity, allowing for one-step purifications of low expressing recombinant proteins.
From the laboratory of Thomas U. Schwartz, PhD, Massachusetts Institute of Technology.
| Catalog Number | Product | DataSheet | Size | AVAILABILITY | Price | Qty |
|---|
Notice to Buyer/User: The buyer/user has a non-exclusive license to use this product for research purposes only. For commercial use of this product, both non-profit and for-profit buyers and users should contact us to inquire about a license from New England Biolabs, Inc.
| Product Type: | Bacteria |
| Name: | LOBSTR E. coli Expression Strain |
| Cell Type: | Chemically competent (CaCl2method) |
| Organism: | E. coli BL21(DE3) |
| Competency: | >1x106cfu/ug DNA |
| Growth Conditions: | Standard E. coli Growth Media (LB, SOC, etc.) at 37C |
| Transformation: | Standard heatshock protocol (42C for 20 seconds) |
| Induction: | IPTG up to 1mM |
| Comments: | Derived from E. coli BL21(DE3) |
| Storage: | -80C (avoid freeze-thaw cycles) |
| Shipped: | Dry ice |
LOBSTR and the parental BL21(DE3) strain show comparable growth
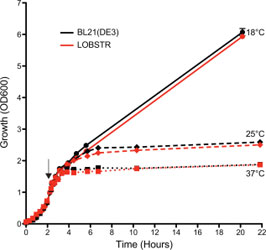
The growth (OD600) of both LOBSTR and the parental BL21(DE3) strain was measured from initial synchronization at 0 h until the final harvest. Both strains carried the same expression plasmid and were grown at 37C until an OD600 ~0.7, at which point protein expression was induced at 18, 25, and 37C (black arrow). The growth curves for LOBSTR and BL21(DE3) are shown in red and black, respectively. Cell growth during log phase and final cell density was similar for both strains. Depending on the expression plasmid different growth behavior was observed, but typically no growth difference was seen between LOBSTR and BL21(DE3).
ArnA and SlyD are eliminated from His-tag purifications from LOBSTR
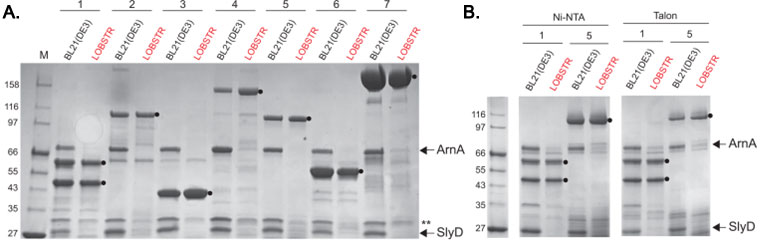
Elution samples of test purifications from BL21(DE3) and LOBSTR using common metal affinity resins are shown. (A) Seven protein constructs were purified from both the parental BL21(DE3) strain and LOBSTR using Ni Sepharose 6FF resin (GE Healthcare). The constructs are numbered 1-7, and contain either a 6×His-tag (1 and 4) or a 10×His-tag (2,3,57). The elution samples were run on an SDS-PAGE gel and stained with Coomassie Blue R250. ArnA and SlyD are indicated by arrows and target proteins indicated with a black circle (). The double asterisk (**) indicates Hsp15, another protein showing reduced Ni-binding affinity in LOBSTR. (B) Purifications of constructs 1 and 5 from BL21(DE3) and LOBSTR were also carried out on two additional commonly used resins, Ni-NTA (Qiagen) and Talon (Clontech). In each case, ArnA and SlyD are successfully eliminated in LOBSTR.
Adapted from: Andersen KR, et al. Proteins. 2013 Nov;81(11):1857-61.
LOBSTR E. Coli strain characterization
- Andersen KR, Leksa NC, Schwartz TU. Optimized E. coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins. 2013 Nov;81(11):1857-61.
LOBSTR E. Coli strain utilization
- Esra Demircioglu F, Cruz VE, Schwartz TU. Purification and Structural Analysis of SUN and KASH Domain Proteins. Methods Enzymol. 2016;569:63-78. View Article
- Kelley K, Knockenhauer KE, Kabachinski G, Schwartz TU. Atomic structure of the Y complex of the nuclear pore. Nat Struct Mol Biol. 2015 May;22(5):425-31. doi: 10.1038/nsmb.2998. Epub 2015 Mar 30.
- Sosa BA, Demircioglu FE, Chen JZ, Ingram J, Ploegh HL, Schwartz TU. How lamina-associated polypeptide 1 (LAP1) activates Torsin. Elife. 2014 Aug 22;3:e03239. doi: 10.7554/eLife.03239.
- Knockenhauer KE, Schwartz TU. Structural Characterization of Bardet-Biedl Syndrome 9 Protein (BBS9). J Biol Chem. 2015 Jun 17. pii: jbc.M115.649202.
- Saxton RA, Knockenhauer KE, Wolfson RL, Chantranupong L, Pacold ME, Wang T, Schwartz TU, Sabatini DM. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science. 2016 Jan 1;351(6268):53-8. doi: 10.1126/science.aad2087. View Article
- Saxton RA, Chantranupong L, Knockenhauer KE, Schwartz TU, Sabatini DM. Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature. 2016 Aug 11;536(7615):229-33. View Article
- Huhn AJ, Guerra RM, Harvey EP, Bird GH, Walensky LD. Selective CovalentTargeting of Anti-Apoptotic BFL-1 by Cysteine-Reactive Stapled PeptideInhibitors. Cell Chem Biol. 2016 Sep 7. pii: S2451-9456(16)30289-6. View Article
- Demircioglu FE, Sosa BA, Ingram J, Ploegh HL, Schwartz TU. Structures ofTorsinA and its disease-mutant complexed with an activator reveal the molecularbasis for primary dystonia. Elife. 2016 Aug 4;5. pii: e17983. View Article
- Truttmann MC, Cruz VE, Guo X, Engert C, Schwartz TU, Ploegh HL. The Caenorhabditis elegans Protein FIC-1 Is an AMPylase That Covalently Modifies Heat-Shock 70 Family Proteins, Translation Elongation Factors and Histones. PLoS Genet. 2016 May 3;12(5):e1006023. View Article
- Saxton RA, Knockenhauer KE, Schwartz TU, Sabatini DM. The apo-structure of the leucine sensor Sestrin2 is still elusive. Sci Signal. 2016 Sep 20;9(446):ra92. doi: 10.1126/scisignal.aah4497. PubMed PMID: 27649739; PubMed Central PMCID: PMC5087270. View Article
- Lawrence KS, Tapley EC, Cruz VE, Li Q, Aung K, Hart KC, Schwartz TU, Starr DA, Engebrecht J. LINC complexes promote homologous recombination in part through inhibition of nonhomologous end joining. J Cell Biol. 2016 Dec 19;215(6):801-821. Epub 2016 Dec 12. View Article
- Kawaharada Y, Nielsen MW, Kelly S, James EK, Andersen KR, Rasmussen SR, Füchtbauer W, Madsen LH, Heckmann AB, Radutoiu S, Stougaard J. Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nat Commun. 2017 Feb 23;8:14534. doi: 10.1038/ncomms14534. PubMed PMID: 28230048; PubMed Central PMCID: PMC5331223. View Article
- Sander B, Xu W, Eilers M, Popov N, Lorenz S. A conformational switch regulates the ubiquitin ligase HUWE1. Elife. 2017 Feb 14;6. pii: e21036. doi: 10.7554/eLife.21036. PubMed PMID: 28193319; PubMed Central PMCID: PMC5308896. View Article
- Andersen KR. Insights into Rad3 kinase recruitment from the crystal structure of the DNA damage checkpoint protein Rad26. J Biol Chem. 2017 Mar 17. pii: jbc.M117.780189. View Article
- Harvey EP, Seo HS, Guerra RM, Bird GH, Dhe-Paganon S, Walensky LD. Crystal Structures of Anti-apoptotic BFL-1 and Its Complex with a Covalent Stapled Peptide Inhibitor. Structure. 2017 Dec 6. pii: S0969-2126(17)30370-2. View Article
- Chen X, Nomani A, Patel N, Hatefi A. Production of low-expressing recombinant cationic biopolymers with high purity. Protein Expr Purif. 2017 Jun;134:11-17. View Article
- Smith ML, Cui W, Jackobel AJ, Walker-Kopp N, Knutson BA. Reconstitution of RNA Polymerase I Upstream Activating Factor and the Roles of Histones H3 and H4 in Complex Assembly. J Mol Biol. 2018 Jan 29. pii: S0022-2836(18)30006-8. View Article
- Kenney GE, Dassama LMK, Pandelia ME, Gizzi AS, Martinie RJ, Gao P, DeHart CJ, Schachner LF, Skinner OS, Ro SY, Zhu X, Sadek M, Thomas PM, Almo SC, Bollinger JM Jr., Krebs C, Kelleher NL, Rosenzweig AC. The biosynthesis of methanobactin. Science. 2018 Mar 23;359(6382):1411-1416. View Article
- Pillon MC, Sobhany M, Borgnia MJ, Williams JG, Stanley RE. Grc3 programs the essential endoribonuclease Las1 for specific RNA cleavage. Proc Natl Acad Sci U S A. 2017 Jul 11;114(28):E5530-E5538. View Article
- Chen X, Nomani A, Patel N, Nouri FS, Hatefi A. Bioengineering a non-genotoxic vector for genetic modification of mesenchymal stem cells. Biomaterials. 2018 Jan;152:1-14. View Article
- Hackenberg C, Hakanpää J, Cai F, Antonyuk S, Eigner C, Meissner S, Laitaoja M, Jänis J, Kerfeld CA, Dittmann E, Lamzin VS. Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria. Proc Natl Acad Sci U S A. 2018 Jun 18. pii: 201806668. View Article
- Montón Silva A, Otten C, Biboy J, Breukink E, VanNieuwenhze M, Vollmer W, den Blaauwen T. The Fluorescent D-Amino Acid NADA as a Tool to Study the Conditional Activity of Transpeptidases in Escherichia coli. Front Microbiol. 2018 Sep 4;9:2101.View Article
- Guerra RM, Bird GH, Harvey EP, Dharia NV, Korshavn KJ, Prew MS, Stegmaier K, Walensky LD. Precision Targeting of BFL-1/A1 and an ATM Co-dependency in Human Cancer. Cell Rep. 2018 Sep 25;24(13):3393-3403.e5. View Article
- Nemec AA, Peterson AK, Warnock JL, Reed RG, Tomko RJ Jr. An Allosteric Interaction Network Promotes Conformation State-Dependent Eviction of the Nas6 Assembly Chaperone from Nascent 26S Proteasomes. Cell Rep. 2019 Jan 8;26(2):483-495.e5. View Article
- Nanji T, Liu X, Chew LH, Li FK, Biswas M, Yu ZQ, Lu S, Dong MQ, Du LL, Klionsky DJ, Yip CK. Conserved and unique features of the fission yeast core Atg1 complex. Autophagy. 2017;13(12):2018-2027. View Article
- Onischenko E, Tang JH, Andersen KR, Knockenhauer KE, Vallotton P, Derrer CP, Kralt A, Mugler CF, Chan LY, Schwartz TU, Weis K. Natively Unfolded FG Repeats Stabilize the Structure of the Nuclear Pore Complex. Cell. 2017 Nov 2;171(4):904-917.e19. View Article
- Ye Q, Kim DH, Dereli I, Rosenberg SC, Hagemann G, Herzog F, Tóth A, Cleveland DW, Corbett KD. The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding. EMBO J. 2017 Aug 15;36(16):2419-2434. View Article
- Wiebach V, Mainz A, Siegert MJ, Jungmann NA, Lesquame G, Tirat S, Dreux-Zigha A, Aszodi J, Le Beller D, Süssmuth RD. The anti-staphylococcal lipolanthines are ribosomally synthesized lipopeptides. Nat Chem Biol. 2018 Jul;14(7):652-654. View Article
- Cruz VE, Schwartz TU. Recombinant Purification of the Periplasmic Portion of the LINC Complex. Methods Mol Biol. 2018;1840:17-23. View Article
- Demircioglu FE, Zheng W, McQuown AJ, Maier NK, Watson N, Cheeseman IM, Denic V, Egelman EH, Schwartz TU. The AAA + ATPase TorsinA polymerizes into hollow helical tubes with 8.5 subunits per turn. Nat Commun. 2019 Jul 22;10(1):3262. View Article
- Bilokapic S, Halic M. Nucleosome and ubiquitin position Set2 to methylate H3K36. Nat Commun. 2019;10(1):3795. Published 2019 Aug 22. View Article
- Obradovic M, Pasternak JA, Hon Ng S, Allan B, Brownlie R, Wilson HL. Immunoproteomic analysis of Lawsonia intracellularis identifies candidate neutralizing antibody targets for use in subunit vaccine development. Vet Microbiol. 2019;235:270-279. View article
- Sonn-Segev A, Belacic K, Bodrug T, et al. Quantifying the heterogeneity of macromolecular machines by mass photometry. Nat Commun. 2020;11(1):1772. Published 2020 Apr 14. View article
- Pillon MC, Goslen KH, Gordon J, Wells ML, Williams JG, Stanley RE. It takes two (Las1 HEPN endoribonuclease domains) to cut RNA correctly. J Biol Chem. 2020;295(18):5857-5870. View article
- Draganova EB, Zhang J, Zhou ZH, Heldwein EE. Structural basis for capsid recruitment and coat formation during HSV-1 nuclear egress. Elife. 2020;9:e56627. Published 2020 Jun 24. View article
- Pillon MC, Goslen KH, Gordon J, Wells ML, Williams JG, Stanley RE. It takes two (Las1 HEPN endoribonuclease domains) to cut RNA correctly. J Biol Chem. 2020;295(18):5857-5870. View article
- Cornacchione LP, Hu LT. Hydrogen peroxide-producing pyruvate oxidase from Lactobacillus delbrueckii is catalytically activated by phosphotidylethanolamine. BMC Microbiol. 2020;20(1):128. Published 2020 May 24. View article
- Kim HR, Xu J, Maeda S, et al. Structural mechanism underlying primary and secondary coupling between GPCRs and the Gi/o family. Nat Commun. 2020;11(1):3160. Published 2020 Jun 22. View article
- Cornacchione LP, Hu LT. Hydrogen peroxide-producing pyruvate oxidase from Lactobacillus delbrueckii is catalytically activated by phosphotidylethanolamine. BMC Microbiol. 2020;20(1):128. Published 2020 May 24. doi:10.1186/s12866-020-01788-6. View article
- Peng HM, Valentín-Goyco J, Im SC, et al. Expression in Escherichia Coli, Purification, and Functional Reconstitution of Human Steroid 5α-Reductases. Endocrinology. 2020;161(8):bqaa117. View article
- McRae EKS, Davidson DE, McKenna SA. 2D Saturation Transfer Difference NMR for Determination of Protein Binding Sites on RNA Guanine Quadruplexes. Methods Mol Biol. 2020;2161:101-113. View article
- Cornacchione LP, Klein BA, Duncan MJ, Hu LT. Interspecies Inhibition of Porphyromonas gingivalis by Yogurt-Derived Lactobacillus delbrueckii Requires Active Pyruvate Oxidase. Appl Environ Microbiol. 2019 Aug 29;85(18):e01271-19. View article
- Barski MS, Minnell JJ, Hodakova Z, Pye VE, Nans A, Cherepanov P, Maertens GN. Cryo-EM structure of the deltaretroviral intasome in complex with the PP2A regulatory subunit B56γ. Nat Commun. 2020 Oct 7;11(1):5043. View article
- Gordon DE, et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020 Dec 4;370(6521):eabe9403. View Article
- Kappenberger J, Koelmel W, Schoenwetter E, Scheuer T, Woerner J, Kuper J, Kisker C. How to limit the speed of a motor: the intricate regulation of the XPB ATPase and translocase in TFIIH. Nucleic Acids Res. 2020 Dec 2;48(21):12282-12296. View article
- Nordeen SA, Turman DL, Schwartz TU. Yeast Nup84-Nup133 complex structure details flexibility and reveals conservation of the membrane anchoring ALPS motif. Nat Commun. 2020 Nov 27;11(1):6060. View article
- Nordeen SA, Andersen KR, Knockenhauer KE, Ingram JR, Ploegh HL, Schwartz TU. A nanobody suite for yeast scaffold nucleoporins provides details of the nuclear pore complex structure. Nat Commun. 2020 Dec 2;11(1):6179. View article
- Lim SM, Cruz VE, Antoku S, Gundersen GG, Schwartz TU. Structures of FHOD1-Nesprin1/2 complexes reveal alternate binding modes for the FH3 domain of formins. Structure. 2021 Jan 19:S0969-2126(20)30480-9. View article
- Lu S, Ye Q, Singh D, Cao Y, Diedrich JK, Yates JR 3rd, Villa E, Cleveland DW, Corbett KD. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat Commun. 2021 Jan 21;12(1):502. View article
- Draganova EB, Heldwein EE. Virus-derived peptide inhibitors of the herpes simplex virus type 1 nuclear egress complex. Sci Rep. 2021 Feb 18;11(1):4206. View article
- Rajavel M, Kumar V, Nguyen H, Wyatt J, Marshall SH, Papp-Wallace KM, Deshpande P, Bhavsar S, Yeole R, Bhagwat S, Patel M, Bonomo RA, van den Akker F. Structural Characterization of Diazabicyclooctane β-Lactam "Enhancers" in Complex with Penicillin-Binding Proteins PBP2 and PBP3 of Pseudomonas aeruginosa. mBio. 2021 Feb 16;12(1):e03058-20. View article
- Gurnani Serrano CK, Winkle M, Martorana AM, Biboy J, Morè N, Moynihan P, Banzhaf M, Vollmer W, Polissi A. ActS activates peptidoglycan amidases during outer membrane stress in Escherichia coli. Mol Microbiol. 2021 Mar 4. View article
If you publish research with this product, please let us know so we can cite your paper.


