Free Methionine-(R)-Sulfoxide Reductase (fRMsr), (C84S, C94S)
Free Methionine-(R)-Sulfoxide Reductase (C84S, C94S), or fRMsr, can be selectively derivatized at a single Cys residue with a variety of Cys-SOH specific probes and be used as a positive control.
Features:
- Reliable procedural and loading control for experiments studying protein cysteine oxidation
- C84S, C94S double mutant allows for 100% sulfenic acid (-SOH) form following treatment with methionine sulfoxide
- Incorporation rates are unaffected by changes in buffer pH (pH 5.5-8.0 tested)
- Compatible with all cysteine oxidation probes
The Free Methionine-(R)-Sulfoxide Reductase (fRMsr) reduces free methionine sulfoxide (Met(O)) to methionine using thiol-disulfide exchange chemistry. This enzyme is involved in oxidative defense and known to form a sulfenic acid intermediate at the active site Cys during the course of turnover. In this variant, all Cys other than the peroxide-sensitive Cys have been removed by mutagenesis in order to stabilize the active site sulfenic acid with respect to disulfide bond formation.
Redox-sensitive cysteine residues in proteins may serve as important components of oxidative signaling or sensors of oxidative stress. Cysteine sulfenic acid modification is an emerging area of interest for those studying biological signal transduction within the cell.
From the laboratory of W. Todd Lowther, PhD, Wake Forest School of Medicine.
 Part of The Investigator's Annexe program.
Part of The Investigator's Annexe program.
Free Methionine-(R)-Sulfoxide Reductase (C84S, C94S), or fRMsr, can be selectively derivatized at a single Cys residue with a variety of Cys-SOH specific probes and be used as a positive control.
Features:
- Reliable procedural and loading control for experiments studying protein cysteine oxidation
- C84S, C94S double mutant allows for 100% sulfenic acid (-SOH) form following treatment with methionine sulfoxide
- Incorporation rates are unaffected by changes in buffer pH (pH 5.5-8.0 tested)
- Compatible with all cysteine oxidation probes
The Free Methionine-(R)-Sulfoxide Reductase (fRMsr) reduces free methionine sulfoxide (Met(O)) to methionine using thiol-disulfide exchange chemistry. This enzyme is involved in oxidative defense and known to form a sulfenic acid intermediate at the active site Cys during the course of turnover. In this variant, all Cys other than the peroxide-sensitive Cys have been removed by mutagenesis in order to stabilize the active site sulfenic acid with respect to disulfide bond formation.
Redox-sensitive cysteine residues in proteins may serve as important components of oxidative signaling or sensors of oxidative stress. Cysteine sulfenic acid modification is an emerging area of interest for those studying biological signal transduction within the cell.
From the laboratory of W. Todd Lowther, PhD, Wake Forest School of Medicine.
 Part of The Investigator's Annexe program.
Part of The Investigator's Annexe program.
| Catalog Number | Product | DataSheet | Size | AVAILABILITY | Price | Qty |
|---|
| Product Type: | Protein |
| Accession ID: | P76270, 946086 |
| Source: | E. Coli |
| Molecular Weight: | 18,752 Da |
| Amino Acid Sequence: | (gidpft)*mnktefyadlnrdfnalmagetsflatlantsallyerltdinwagfylleddtlvlgpfqgkia(c>s)vripvgrgv(c>s)gtavarnqvqriedvhvfdghiacdaasnseivlplvvknqiigvldidstvfgrftdedeqglrqlvaqlekvlattdykkffasvag (* these residues remain following the cleavage of the His-tag) |
| Purity: | >98% by SDS-PAGE |
| Buffer: | 20mM Hepes pH 7.5, 100mM NaCl |
| Concentration: | 10mg/mL |
| Comments: | C84S, C94S |
| Storage: | -80C |
| Shipped: | Dry ice |
SDS-PAGE and MALDI-TOF Mass Spectrometry Analyses of Free Methionine-(R)-Sulfoxide Reductase (fRMsr), (C84S, C94S)
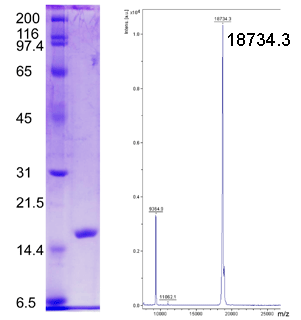
(Left) SDS-PAGE Analysis. Protein ladder (kDa) on left and purified fRMsr on right. (Right) MALDI-TOF analysis of purified fRMsr.
For experiments, the Cys sulfenic acid form of fRMsr should be prepared in advance by incubation with a 100-fold excess of methionine sulfoxide for 2 min and removal of the excess amino acid using a Bio-Gel P6 column. The protein can then be diluted into a solution containing 2 mM DCP-Bio1 (or other cysteine oxidation probe) to give final concentrations of 50 uM fRMsr and 1 mM DCP-Bio1 in 5 mM potassium phosphate buffer, pH 7.0 Mutant fRMsr can also be biotinylated with biotin maleimide (see Nelson et al., 2010, for detailed protocol) and 1 ug fRMsr/500 ug of cell lysate added to each sample prior to affinity capture for use as a procedural and loading control.
- Lin Z, Johnson LC, Weissbach H, Brot N, Lively MO, and Lowther WT. Free methionine-(R)-sulfoxide reductase from Escherichia coli reveals a new GAF domain function. PNAS 104(23): 9597-9602 (2007).
- Klomsiri C, Nelson KJ, Bechtold E, Soito L, Johnson LC, Lowther WT, Ryu S, King SB, Furdui CM and Poole LB. Use of dimedone-based chemical probes for sulfenic acid detection; evaluation of conditions affecting probe incorporation into redox sensitive proteins. Methods. Enzymol. 473: 77-94. (2010).
- Nelson, K.J., Klomsiri, C., Codreanu, S.G., Soito, L., Liebler, D.C., Rogers, L.C., Daniel, L.W. & Poole, L.B. Use of dimedone-based chemical probes for sulfenic acid detection; methods to visualize and identify labeled proteins. Methods Enzymol 473, 95-115 (2010).
If you publish research with this product, please let us know so we can cite your paper.


